Hello! I am a computational biophysicist with experience in minimal method development, elastic rods, liquid-liquid phase separation, research software engineering and high-performance computing. Previous: FFEA Group at Leeds, Collepardo Lab at Cambridge. Current: computational scientist at the STFC. You can get in touch with me at [email protected]. While you're here, please take a look at my CV and LinkedIn profile!
My research interests include kirchoff rods, molecular dynamics, and coarse-grained simulations at the molecular mesoscale. I have experience in high-performance computing, development and design of simulations, molecular dynamics, mathematical logic, computational biophysics, photonics and optics, and statistical mechanics.
Physics, Software KOBRA: Fluctuating Elastic Rods for Slender Biological Macromolecules
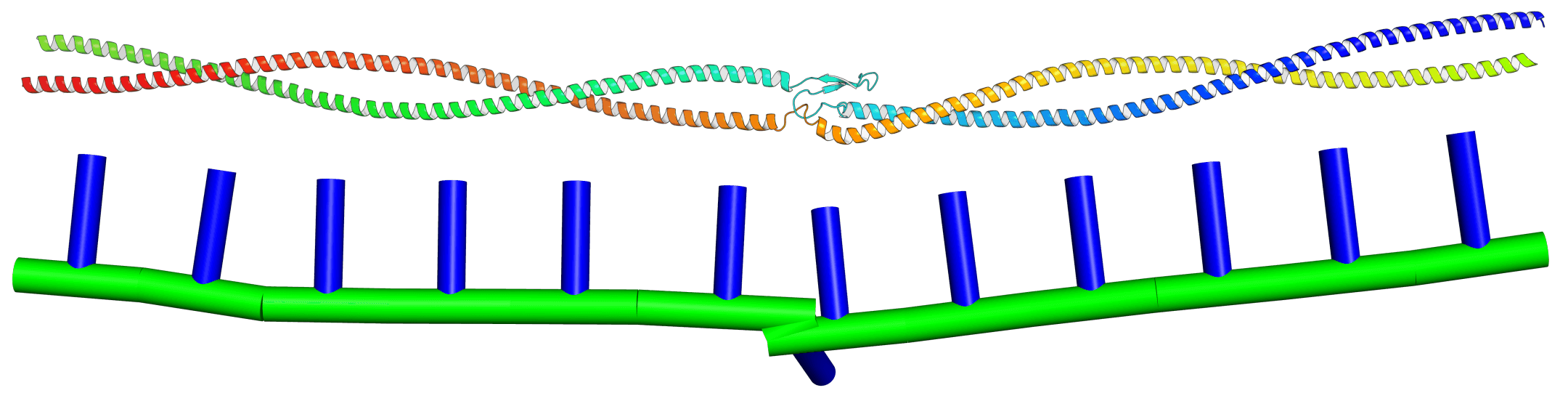 KOBRA is a fluctuating Kirchoff elastic rod algorithm for performing dynamical simulations of slender biomolecules, such as alpha-helices and coiled-coils. It can support parameterisations of objects with arbitrary equilibrium shapes and inhomogeneous, anisotropic material parameters, and it can reach second-scale trajectories of quite large systems on modest workstation hardware. It's written in C++ and is released, for free, under the GPLv3 free software license. So far, KOBRA has been used to build models of Ndc80C, the SMC complex and Myosin-V, and may soon be deployed to study COVID-19 spike proteins. This page will contain links to the KOBRA publication, my PhD thesis, download links and installation instructions.
KOBRA is a fluctuating Kirchoff elastic rod algorithm for performing dynamical simulations of slender biomolecules, such as alpha-helices and coiled-coils. It can support parameterisations of objects with arbitrary equilibrium shapes and inhomogeneous, anisotropic material parameters, and it can reach second-scale trajectories of quite large systems on modest workstation hardware. It's written in C++ and is released, for free, under the GPLv3 free software license. So far, KOBRA has been used to build models of Ndc80C, the SMC complex and Myosin-V, and may soon be deployed to study COVID-19 spike proteins. This page will contain links to the KOBRA publication, my PhD thesis, download links and installation instructions.
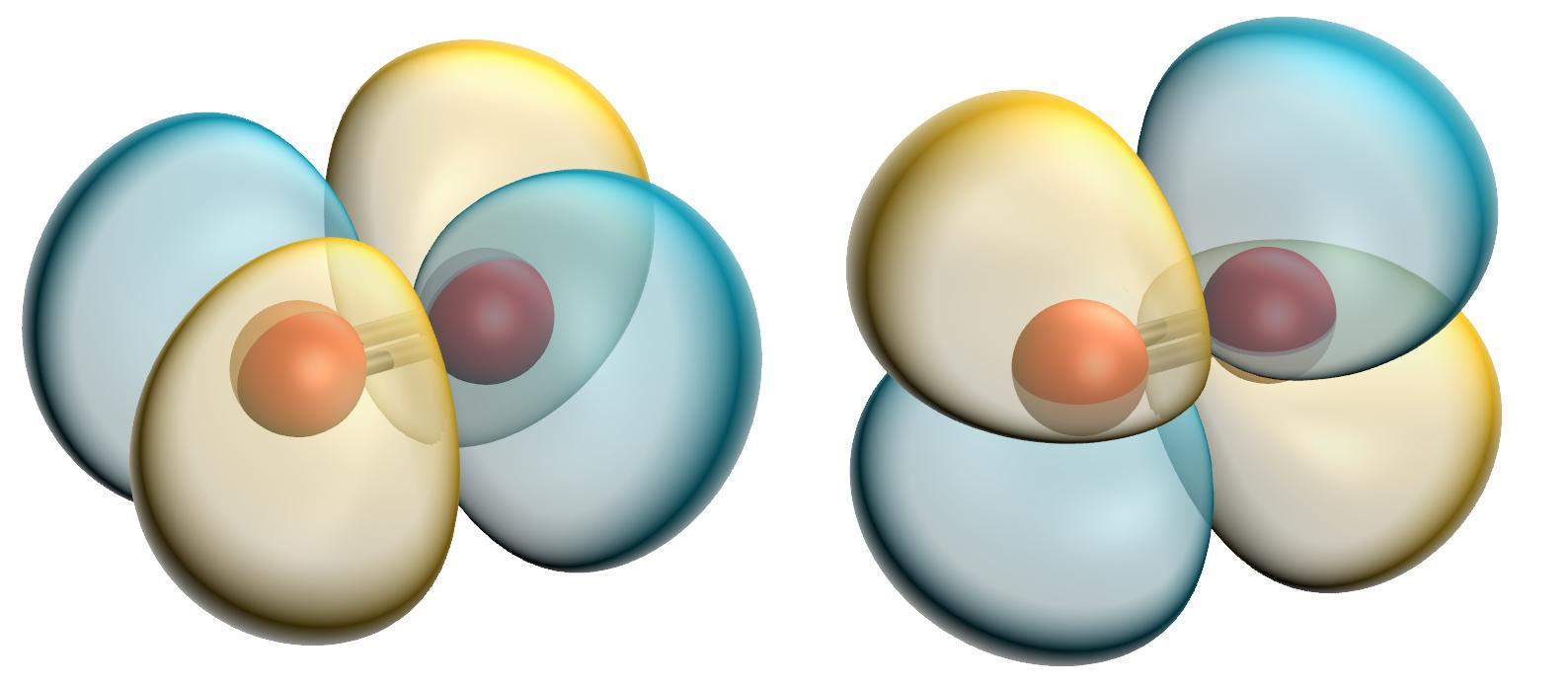 During 2021 I worked as a senior research software engineer on the MOLPRO, quantum chemistry package. This post details my work on optimising and profiling MOLPRO's linear algebra libraries, and the results.
During 2021 I worked as a senior research software engineer on the MOLPRO, quantum chemistry package. This post details my work on optimising and profiling MOLPRO's linear algebra libraries, and the results.  KOBRA is a fluctuating Kirchoff elastic rod algorithm for performing dynamical simulations of slender biomolecules, such as alpha-helices and coiled-coils. It can support parameterisations of objects with arbitrary equilibrium shapes and inhomogeneous, anisotropic material parameters, and it can reach second-scale trajectories of quite large systems on modest workstation hardware. It's written in C++ and is released, for free, under the GPLv3 free software license. So far, KOBRA has been used to build models of Ndc80C, the SMC complex and Myosin-V, and may soon be deployed to study COVID-19 spike proteins. This page will contain links to the
KOBRA is a fluctuating Kirchoff elastic rod algorithm for performing dynamical simulations of slender biomolecules, such as alpha-helices and coiled-coils. It can support parameterisations of objects with arbitrary equilibrium shapes and inhomogeneous, anisotropic material parameters, and it can reach second-scale trajectories of quite large systems on modest workstation hardware. It's written in C++ and is released, for free, under the GPLv3 free software license. So far, KOBRA has been used to build models of Ndc80C, the SMC complex and Myosin-V, and may soon be deployed to study COVID-19 spike proteins. This page will contain links to the 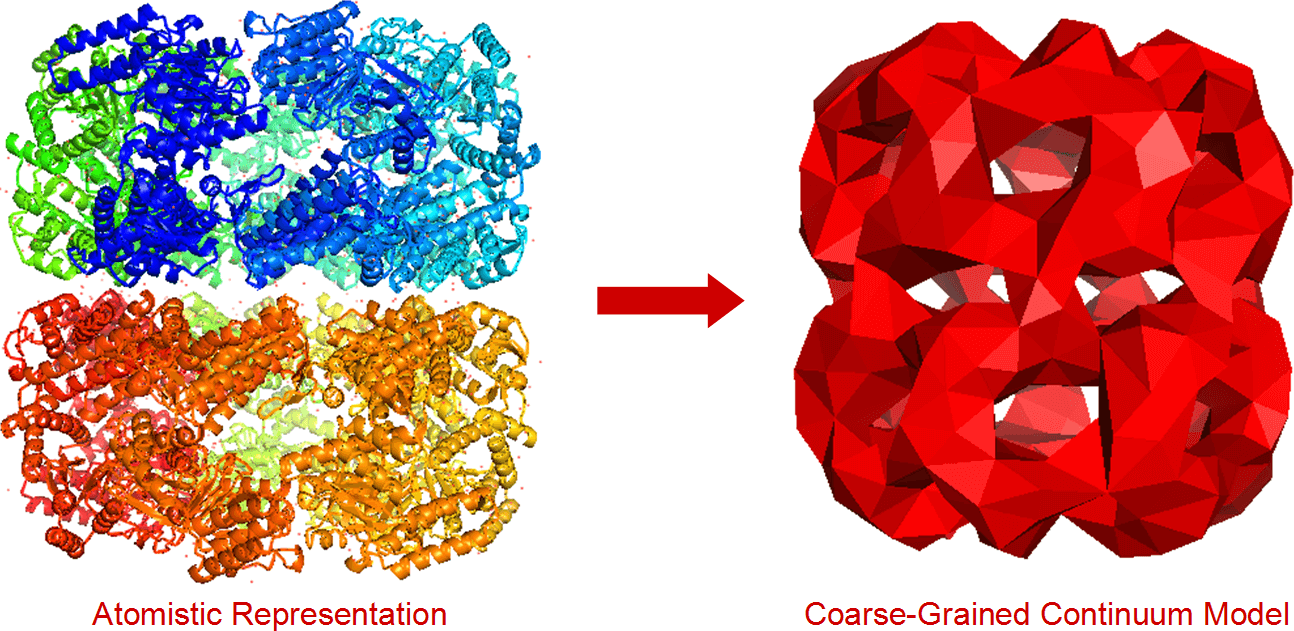
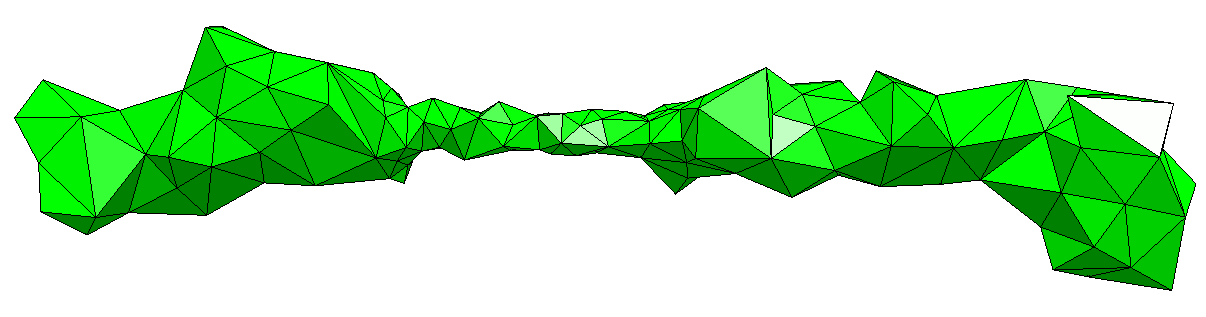 Myosin-VII is part of the Myosin family of motor proteins. It has a key role in the operation of biological structures in the eye and inner ear. Unfortunately, the dynamics of Myosin-VII and its inhibition are poorly-understood. This article details my use of FFEA (Fluctuating Finite Element Analysis), a coarse-grained continuum simulation package, to model the motion of Myosin-VII, and understand its inhibition behaviour.
Myosin-VII is part of the Myosin family of motor proteins. It has a key role in the operation of biological structures in the eye and inner ear. Unfortunately, the dynamics of Myosin-VII and its inhibition are poorly-understood. This article details my use of FFEA (Fluctuating Finite Element Analysis), a coarse-grained continuum simulation package, to model the motion of Myosin-VII, and understand its inhibition behaviour. 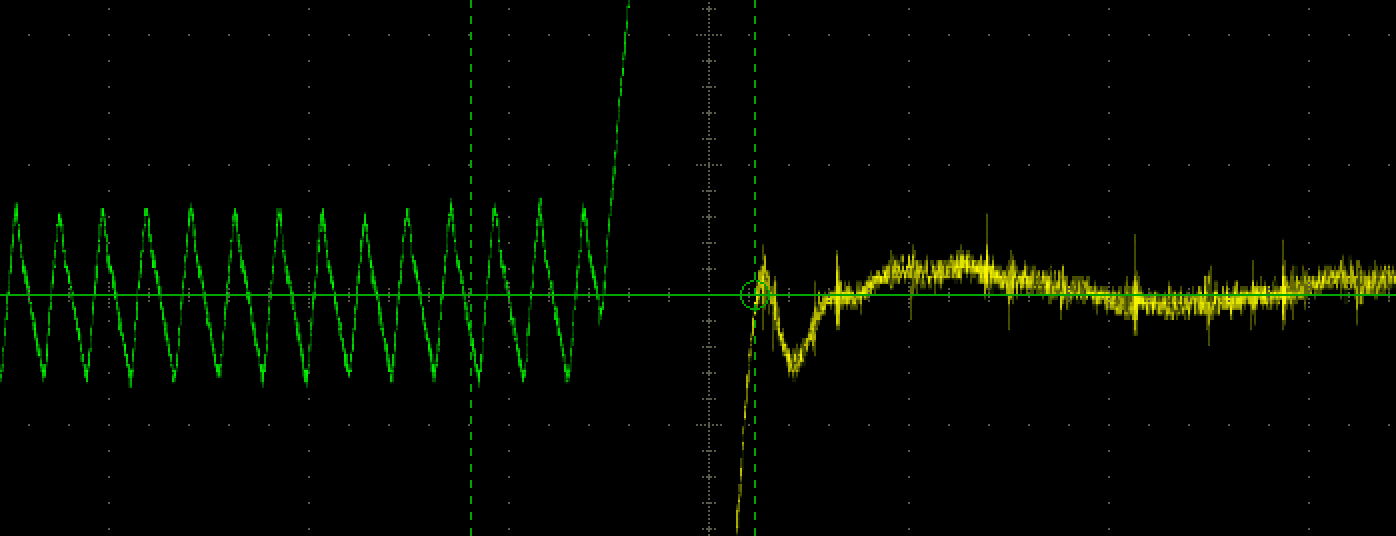 As you probably know, halogen light bulbs are incandescent: they produce light as a side effect of being heated up. Conversely, LEDs are semiconductor devices, and prolonged heat can be very damaging to them. LED devices tend to be cooled in a similar way to computer CPUs - using a thermal interface (like thermal paste) a heatsink, and sometimes a fan as well. However, the effectiveness of these cooling methods can vary wildly, depending on the operational limits of the LED and the type of thermal interface being used. In this article, I'll discuss the experimental methods I designed to characterise the thermal performance of LEDs using a variety of cooling methods.
As you probably know, halogen light bulbs are incandescent: they produce light as a side effect of being heated up. Conversely, LEDs are semiconductor devices, and prolonged heat can be very damaging to them. LED devices tend to be cooled in a similar way to computer CPUs - using a thermal interface (like thermal paste) a heatsink, and sometimes a fan as well. However, the effectiveness of these cooling methods can vary wildly, depending on the operational limits of the LED and the type of thermal interface being used. In this article, I'll discuss the experimental methods I designed to characterise the thermal performance of LEDs using a variety of cooling methods. 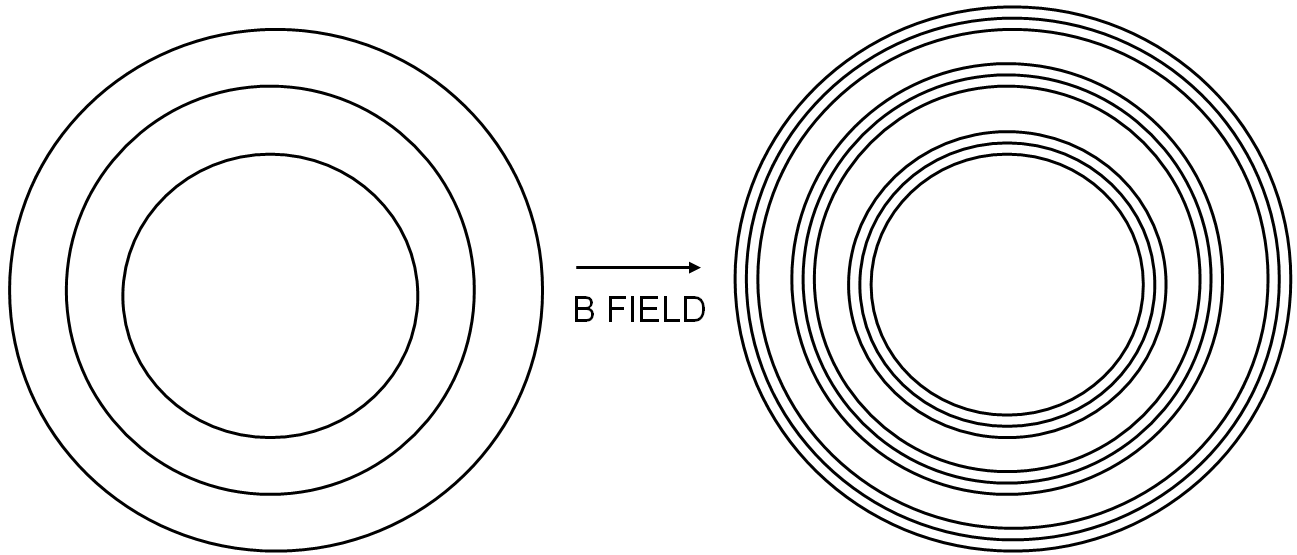 The Bohr Magneton is a physical constant which is used to express the dipole moment of electrons. It corresponds to the angular momentum of an electron in the lowest orbital. The Bohr Magneton relates the splitting of atomic energy levels to the strength of an applied magnetic field. The Zeeman effect offers us an easy way to observe this splitting. In this lab report, I discuss a how to observe the Zeeman effect, and how it can be used to compute an accurate value for the Bohr Magneton.
The Bohr Magneton is a physical constant which is used to express the dipole moment of electrons. It corresponds to the angular momentum of an electron in the lowest orbital. The Bohr Magneton relates the splitting of atomic energy levels to the strength of an applied magnetic field. The Zeeman effect offers us an easy way to observe this splitting. In this lab report, I discuss a how to observe the Zeeman effect, and how it can be used to compute an accurate value for the Bohr Magneton. 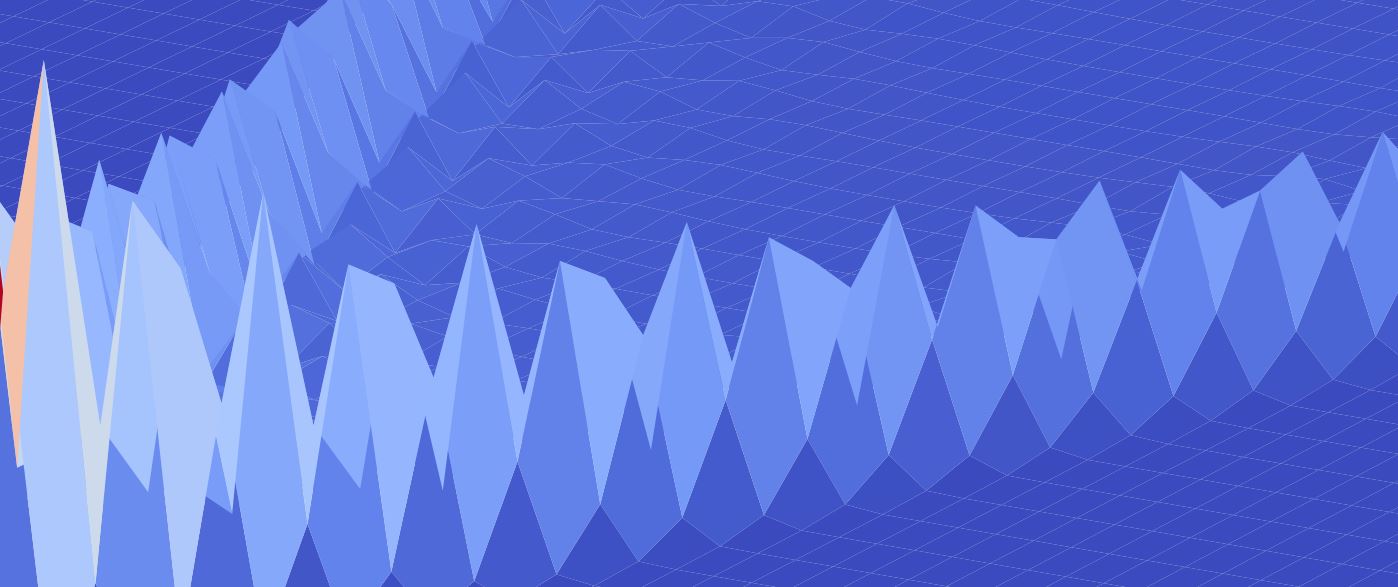 A random walk (sometimes called 'the drunkard's walk' describes the motion of a particle that undergoes a series of random steps. But what happens if that particle is a quantum particle? This article presents a few interesting ways of visualising this motion, using Python and matplotlib.
A random walk (sometimes called 'the drunkard's walk' describes the motion of a particle that undergoes a series of random steps. But what happens if that particle is a quantum particle? This article presents a few interesting ways of visualising this motion, using Python and matplotlib.