KOBRA: Fluctuating Elastic Rods for Slender Biological Macromolecules
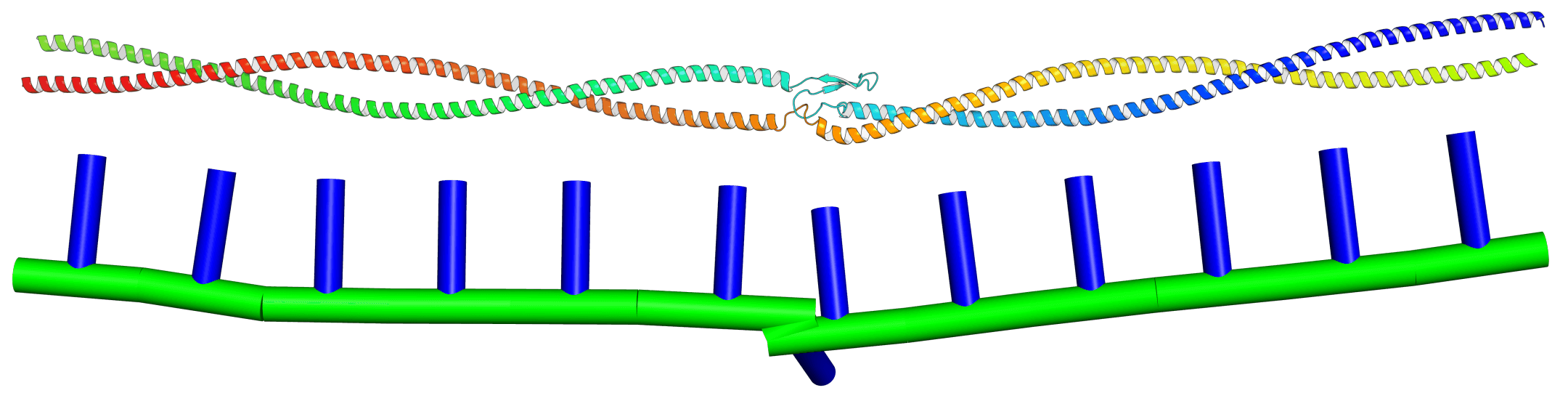
KOBRA is a fluctuating Kirchoff elastic rod algorithm for performing dynamical simulations of slender biomolecules, such as alpha-helices and coiled-coils. It has built-in parameterisation tools, it can support parameterisations of objects with arbitrary equilibrium shapes and inhomogeneous, anisotropic material parameters, and it can reach second-scale trajectories of quite large systems on modest workstation hardware. It’s written in C++ and you can download it for free as part of FFEA, with source code released under the GPLv3 free software license. So far, KOBRA has been used to build models of Ndc80C, the SMC complex and Myosin-V, and may soon be deployed to study COVID-19 spike proteins.
Features
- Coarse-grained kirchoff rod algorithm with three degrees of freedom (stretching, twisting and bending)
- Thermodynamically-correct thermal noise
- Highly flexible parameterisation and parameter recovery from all-atom simulations, can represent molecular hinges
- Exchange of forces with FFEA objects
- Free and open source (GPLv3) with a Python API
- PyMOL extension for visualisation
- Extensive unit and integration tests
Download
The easiest way to download KOBRA is as part of FFEA. Download FFEA+KOBRA as a binary distribution or build from source, following the installation instructions. Then, head over to the KOBRA tutorial to get started. You’ll need some experience in Linux and Python.
Tech
- RNGStreams
- Boost (>=1.54.0)
- MDAnalysis
- PyMOL (>=1.8) – PyMOL 1.8 is supported better
More
You can find a talk I gave on KOBRA here. You can read my PhD thesis on the White Rose repository.
Credits
KOBRA was developed by me, and designed by myself, Daniel Read, Sarah Harris and Oliver Harlen. I would also like to thank Albert Solernou for his HPC advice, and Samantha Coffey for testing.
Last updated January 10, 2022.